Tzu-Tang Lin
AI × Bioinformatics R&D
Motivated to tap into the potential of AI and data science to reveal game-changing capabilities that accelerate the advancement of precision medicine and maximally benefit public health.
Motivated to tap into the potential of AI and data science to reveal game-changing capabilities that accelerate the advancement of precision medicine and maximally benefit public health.


AI × Bioinformatics researcher & developer with extensive cross-disciplinary R&D experience spanning drug discovery, genomics, and biomedical AI. Passionate about transforming complex biological data into actionable insights for advancing precision medicine and improving public health outcomes.
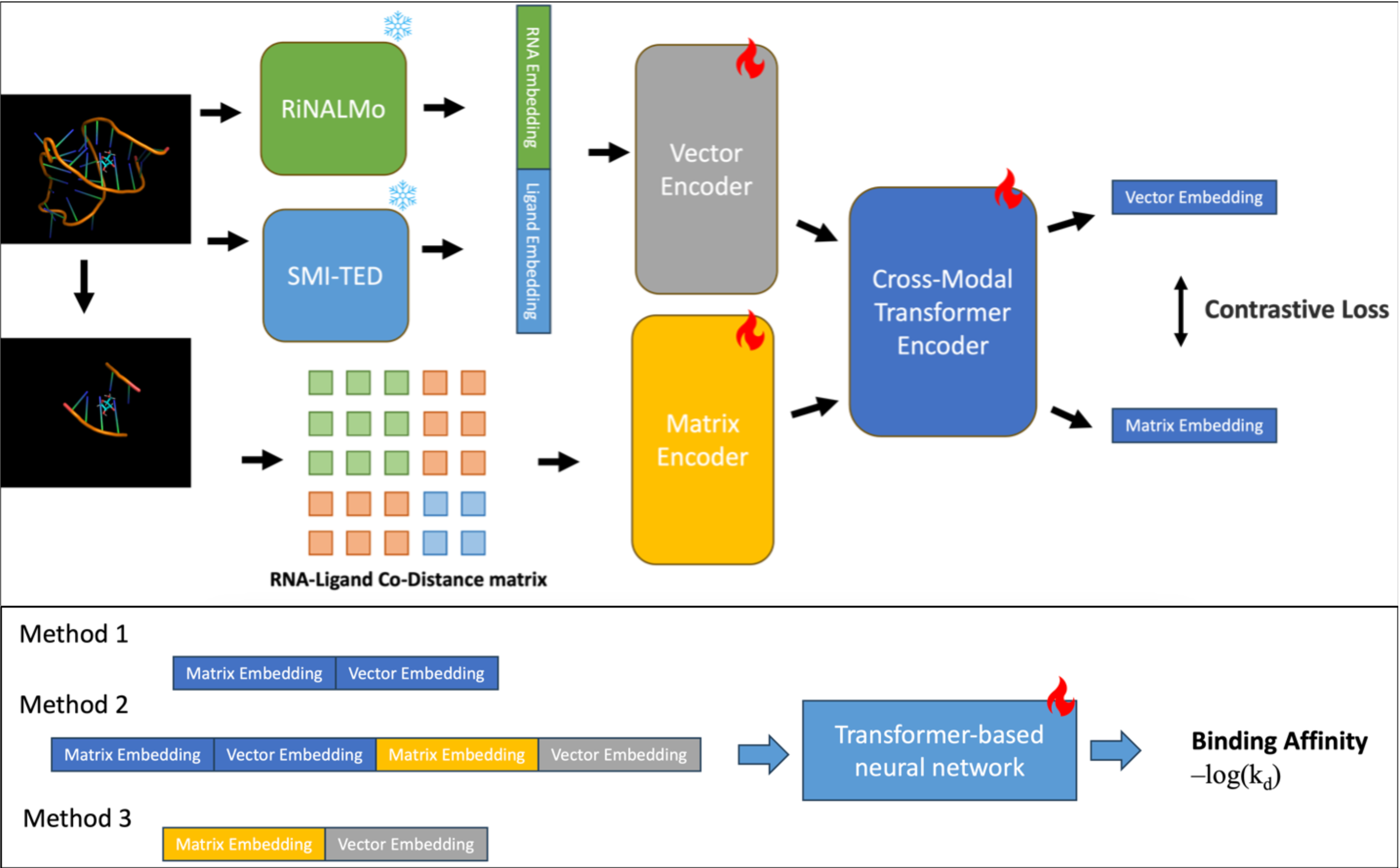
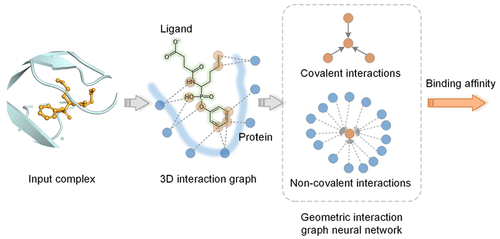
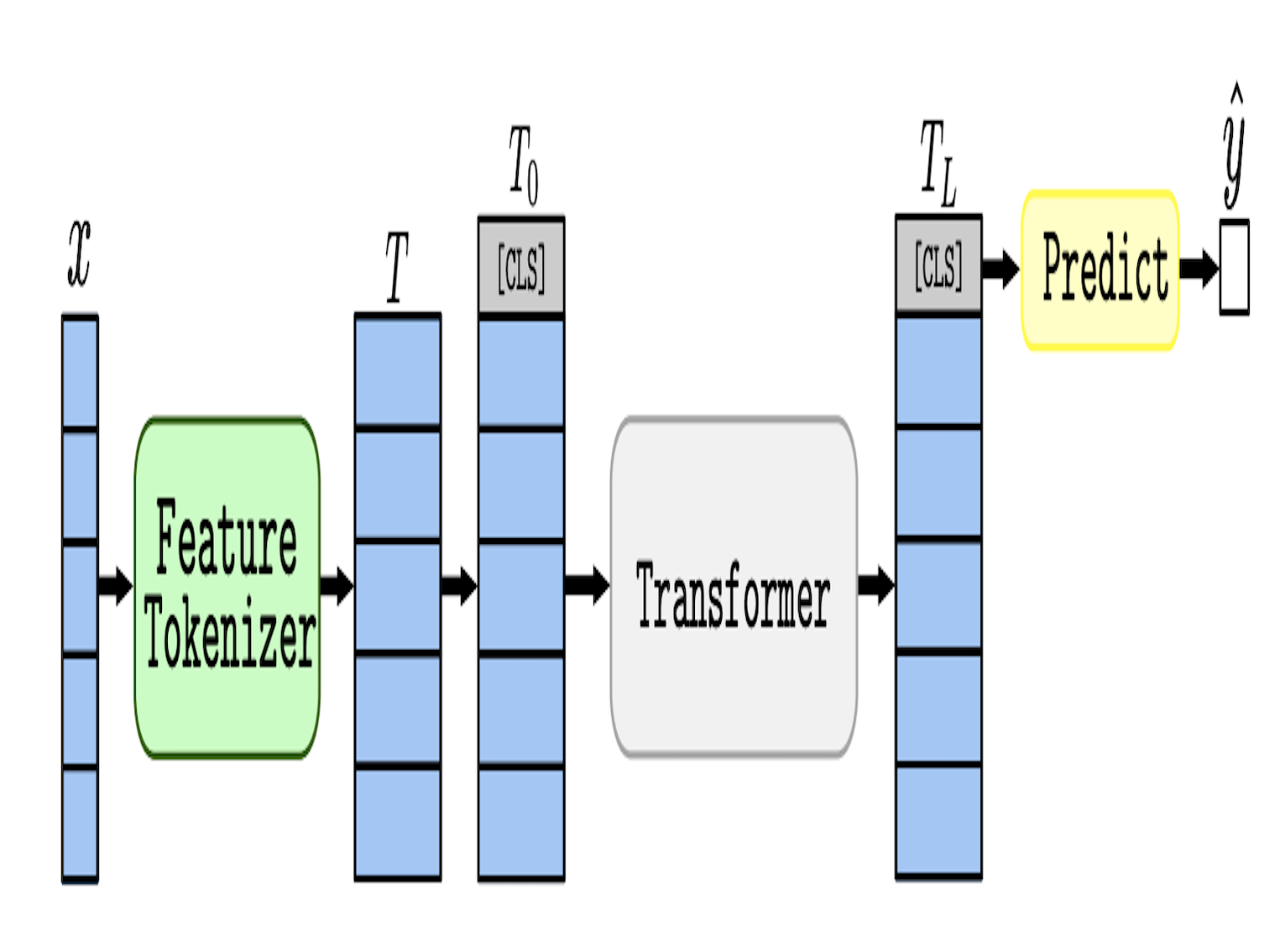
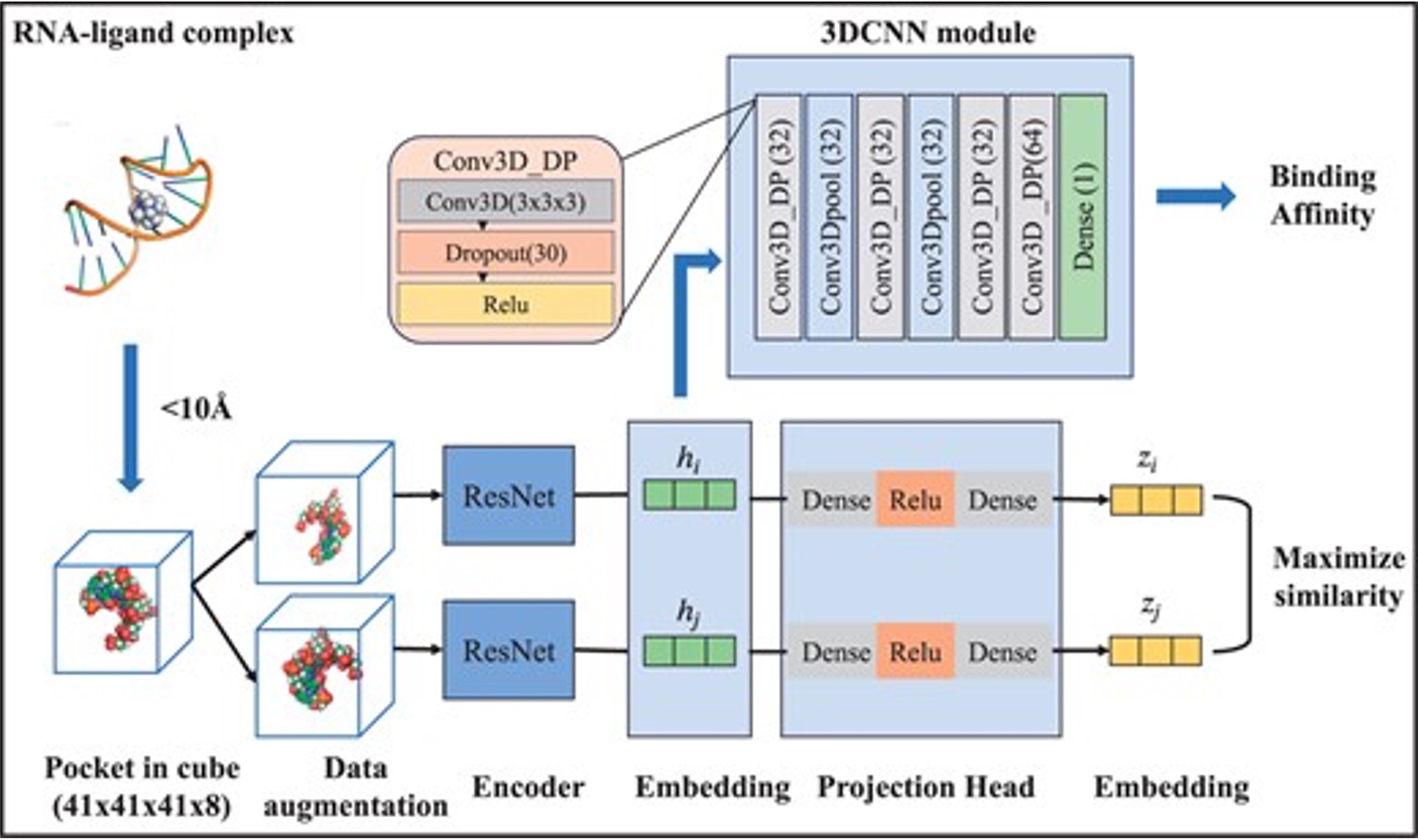
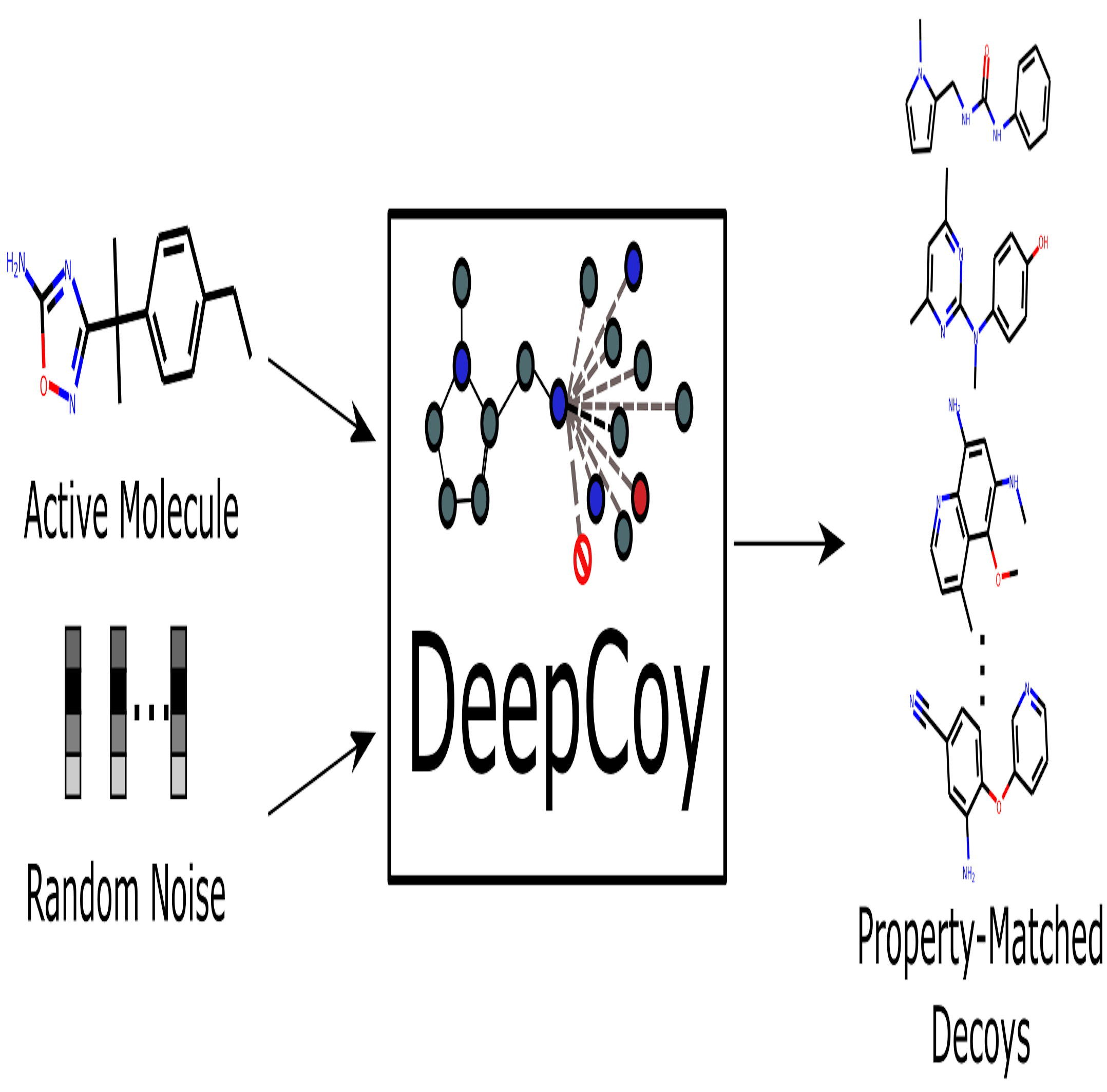
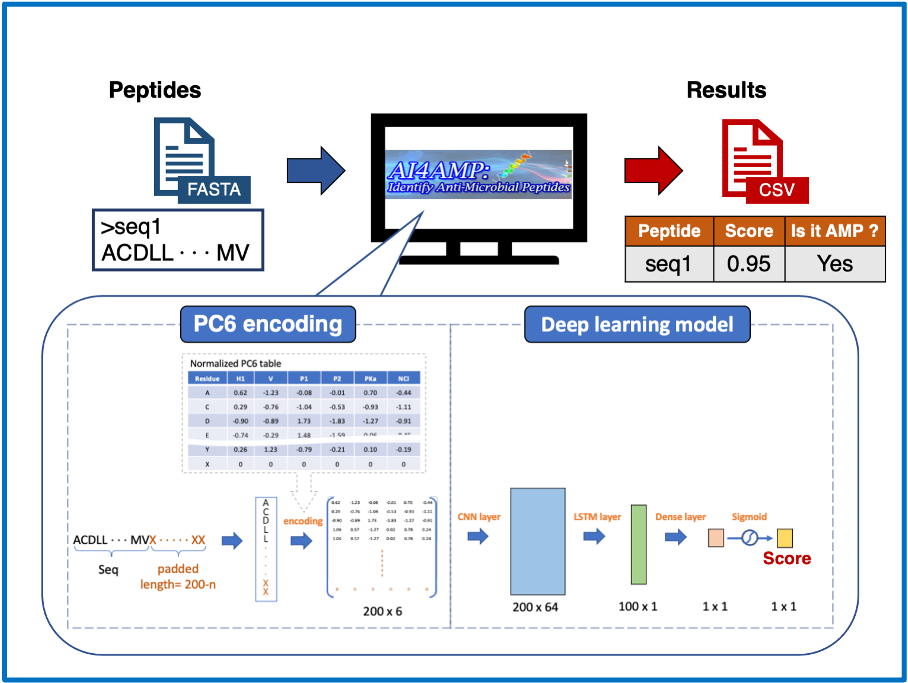
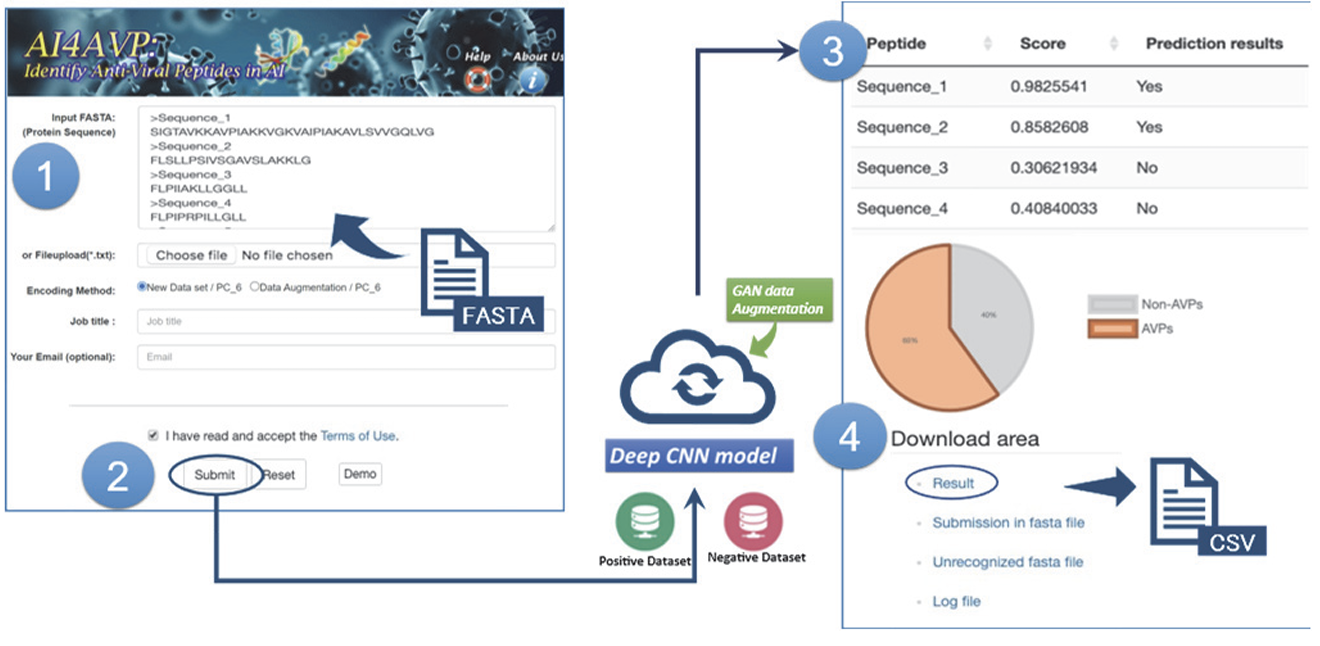
Proven expertise in AI-driven RNA–small molecule drug modeling, antimicrobial peptide and functional protein design, next-generation sequencing (NGS) and genome-wide association studies (GWAS), as well as electronic health records (EHR)–based medical applications.
Experienced in independent research, publishing peer-reviewed articles, serving as a scientific reviewer, and mentoring students. Skilled in leading end-to-end research pipelines—from dataset curation to deep learning model development—and collaborating effectively with academia, hospitals, and industry partners.
Research InterestsSeeking full-time opportunities in AI-driven bioinformatics, drug discovery, and precision medicine.